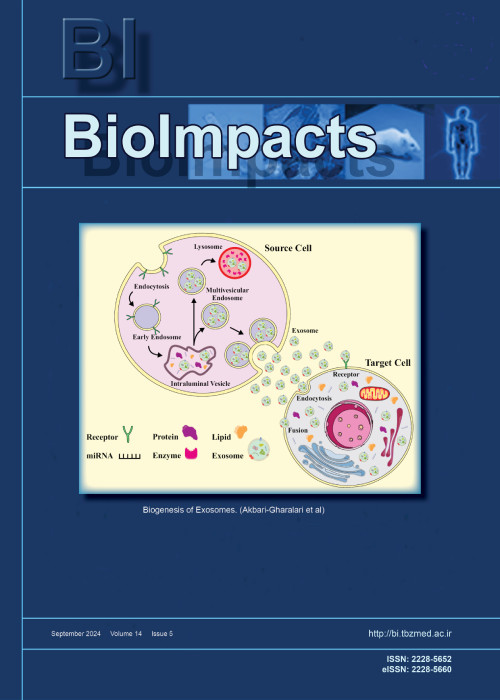
فهرست مطالب
Biolmpacts
Volume:12 Issue: 2, Feb 2022
- تاریخ انتشار: 1401/01/08
- تعداد عناوین: 10
-
-
Pages 87-88
Cancer remains to be a major hurdle to global health. Exosomes as a versatile bio-derived platform, hold a bright prospect in nano-scaled delivery/targeting strategies. Shreds of evidence indicate that exosomes have a critical role in drug resistance in cancer cells through various mechanisms including shuttling of miRNAs, drug efflux transporters, and anti-apoptotic signaling. Exosomes’ cargo, particularly miRNAs, may exert both resistance and in a few cases sensitivity to the anticancer agents in targeted cells. Therefore, the source and components of the exosomes should be carefully considered before any application. Our aim in this editorial is to further highlight the role of exosomes in the development of resistance to therapy in cancer cells. As a new chapter for drug delivery, the challenges should be elucidated before exosomes emerge as novel nanoplatforms for cancer therapy.
Keywords: Cancer therapy, Chemoresistance, Exosomesmi, RNA -
Pages 89-105Introduction
Pompe disease (PD) is a disease caused by pathogenic variations in the GAA gene known as glycogen storage disease type II, characterized by heart hypertrophy, respiratory failure, and muscle hypotonia, leading to premature death if not treated early. The only treatment option, enzyme replacement therapy (ERT), significantly improves the prognosis for some patients while failing to help others. In this study, the determination of key genes involved in the response to ERT and potential molecular mechanisms were investigated.
MethodsGene Expression Omnibus (GEO) data, accession number GSE38680, containing samples of biceps and quadriceps muscles was used. Expression array data were analyzed using BRB-Array Tools. Biceps group patients did not receive ERT, while quadriceps received treatment with rhGAA at 0, 12, and 52 weeks. Differentially expressed genes (DEGs) were deeply analyzed by DAVID, GO, KEGG and STRING online analyses, respectively.
ResultsA total of 1727 genes in the biceps group and 1198 genes in the quadriceps group are expressed differently. It was observed that DEGs were enriched in the group that responded poorly to ERT in the 52nd week. Genes frequently changed in the weak response group; the expression of 530 genes increased and 1245 genes decreased compared to 0 and 12 weeks. The GO analysis demonstrated that the DEGs were mainly involved in vascular smooth muscle contraction, lysosomes, autophagy, regulation of actin cytoskeleton, inflammatory response, and the WNT signaling pathway. We also discovered that the WNT signaling pathway is highly correlated with DEGs. Several DEGs, such as WNT11, WNT5A, CTNNB1, M6PR, MYL12A, VCL, TLN, FYN, YES1, and BCL2, may be important in elucidating the mechanisms underlying poor response to ERT.
ConclusionEarly diagnosis and treatment of PD are very important for the clinic of the disease. As a result, it suggests that the enriched genes and new pathways emerging as a result of the analysis may help identify the group that responds poorly to treatment and the outcome of the treatment. Obtained genes and pathways in neonatal screening will guide diagnosis and treatment.
Keywords: Pompe disease, GEO data, Bioinformatics, Differentially expressed genes, Enzyme replacement treatment, Wnt signaling pathway -
Pages 107-113Introduction
The new species of coronaviruses (CoVs), SARS-CoV-2, was reported as responsible for an outbreak of respiratory disease. Scientists and researchers are endeavoring to develop new approaches for the effective treatment against of the COVID-19 disease. There are no finally targeted antiviral agents able to inhibit the SARS-CoV-2 at present. Therefore, it is of interest to investigate the potential uses of levamisole derivatives, which are reported to be antiviral agents targeting the influenza virus.
MethodsIn the present study, 12 selected levamisole derivatives containing imidazo[2,1-b]thiazole were subjected to molecular docking in order to explore the binding mechanisms between these derivatives and the SARS-CoV-2 Mpro (PDB: 7BQY). The levamisole derivatives were evaluated for in silico ADMET properties for wet-lab applicability. Further, the stability of the best-docked complex was checked using molecular dynamics (MD) simulation at 20 ns.
ResultsLevamisole derivatives and especially molecule N°6 showed more promising docking results, presenting favorable binding interactions as well as better docking energy compared to chloroquine and mefloquine. The results of ADMET prediction and MD simulation support the potential of the molecule N°6 to be further developed as a novel inhibitor able to stop the newly emerged SARS-CoV-2.
ConclusionThis research provided an effective first line in the rapid discovery of drug leads against the novel CoV (SARS-CoV-2).
Keywords: COVID-19, SARS-CoV-2, Levamisole, Molecular docking, Molecular dynamics simulation, In silico ADMET -
Pages 115-126Introduction
Breast cancer is the most serious cause of women’s death throughout the world. Using nanocarrier vehicles to the exact site of cancer upgrades the therapeutic efficiency of the drugs. Capsulation of active proteins in the vesicular liposomes’ hydrophilic core is essential to develop a therapeutic protein carrier system. We aimed to encapsulate the medicinal leech saliva extract (LSE) and assess the inhibition of angiogenesis of breast cancer cells by targeting vascular endothelial growth factor A (VEGFA).
MethodsIn this research, enhanced formulation of liposomal protein was determined by zeta potential analysis, droplet size, drug release assay, and transmission electron microscopy (TEM). Furthermore, a cytotoxicity assay of liposomal LSE was performed to determine the cytotoxic activity of components. For assessing the expression of VEGFA, P53, and hypoxia-inducible factor subunit alpha (HIF1a) genes, Real-Time PCR was applied.
ResultsNano liposome was chosen as an enhanced formulation due to its much smaller size (46.23 nm). Liposomal LSE had more practical actions on the MCF-7 cells. As noticed by DAPI staining, apoptosis was extensively greater in treated MCF-7 cells. Wound healing assay demonstrated that MCF-7 cells could not sustain growth at the presence of liposomal LSE and expression of the VEGFA gene was declined in treated cells. Downregulation of VEGFA was evaluated with western blotting technique.
ConclusionIt can be concluded that our investigation of the tests confirmed the fact that nano liposomal LSE is a novel promising formulation for anticancer drugs and can significantly improve the penetration of protein drugs to cancer cells.
Keywords: Angiogenesis, Breast cancer, Leech saliva extract, Liposome, Nano drug, VEGFA -
Pages 127-138Introduction
Exosomal microRNAs (miRNAs) are emerging diagnostic biomarkers for different types of cancers. We aim to detect gastric cancer (GC)-specific miRNAs in serum exosomes with diagnostic potential.
MethodsA pair of 43 tumor and tumor-adjacent tissue biopsies obtained from GC patients, also 5 mL peripheral blood (following 12h fasting) were collected from the same patients and healthy controls (HCs). QIAGEN miRCURY LNA miRNA Focus PCR Panel applied to screen differentially expressed onco-miRNAs. The candidate miRNAs with the highest fold changes proceeded for validation by qRT-PCR in individuals.
ResultsWe identified that exosomal miR-10a-5p, miR-19b-3p, miR-215-5p, and miR-18a-5p were significantly upregulated in GC patient’s exosomes in contrast to HCs exosomes, Roc curve analysis indicated area under the ROC curve (AUC) of 0.801, 0.721, 0.780 and 0.736 respectively. The Roc curve analysis for the combined signature of four exosomal miRNAs indicated AUC of 0.813. Also, Spearman's correlation coefficients indicated that the miRNA expression is highly correlated between tumor and exosome.
ConclusionHerein, we specifically identified four miRNAs in serum exosomes of GC patients for a diagnostic purpose which are directly associated with tumoral miRNA expression profile.
Keywords: Liquid Biopsy, ExomiR, OncomiR, Stem-loop RT-PCR, Stomach -
Pages 139-146Introduction
With the outbreak of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the interaction between the host and SARS-CoV-2 was widely studied. However, it is unclear whether and how SARS-CoV-2 infection affects lung microflora, which contribute to COVID-19 complications.
MethodsHere, we analyzed the metatranscriptomic data of bronchoalveolar lavage fluid (BALF) of 19 COVID-19 patients and 23 healthy controls from 6 independent projects and detailed the active microbiota landscape in both healthy individuals and COVID-19 patients.
ResultsThe infection of SARS-CoV-2 could deeply change the lung microbiota, evidenced by the α-diversity, β-diversity, and species composition analysis based on bacterial microbiota and virome. Pathogens (e.g., Klebsiella oxytoca causing pneumonia as well), immunomodulatory probiotics (e.g., lactic acid bacteria and Faecalibacterium prausnitzii, a butyrate producer), and Tobacco mosaic virus (TMV) were enriched in the COVID-19 group, suggesting a severe microbiota dysbiosis. The significant correlation between Rothia mucilaginosa, TMV, and SARS-CoV-2 revealed drastic inflammatory battles between the host, SARS-CoV-2, and other microbes in the lungs. Notably, TMV only existed in the COVID-19 group, while human respirovirus 3 (HRV 3) only existed in the healthy group. Our study provides insights into the active microbiota in the lungs of COVID-19 patients and would contribute to the understanding of the infection mechanism of SARS-CoV-2 and the treatment of the disease and complications.
ConclusionSARS-COV-2 infection deeply altered the lung microbiota of COVID-19 patients. The enrichment of several other pathogens, immunomodulatory probiotics (lactic acid or butyrate producers), and TMV in the COVID-19 group suggests a complex and active lung microbiota disorder.
Keywords: Microbiota, SARS-CoV-2, COVID-19, Lactic acid bacteria, Faecalibacterium prausnitzii -
Pages 147-154Introduction
Neurodegenerative diseases are accompanied by loss of neuronal function and integrity. Stem cell therapy is utilized to regenerate neurons to repair the damaged area. Regeneration potential of stem cells can be enhanced by using chemicals with known bioactive properties. In the current study, two bioactive compounds, α-pinene (AP) and thymoquinone (TQ) were explored for their neuronal differentiation potential of rat bone marrow mesenchymal stem cells (MSCs).
MethodsMSCs were isolated, cultured and characterized immunocytochemically for the presence of specific surface markers. Optimized concentrations of both compounds (20 µM AP and 12 µM TQ) as determined by MTT assay, were used to treat MSCs in separate and combined groups. All groups were assessed for the presence of neuronal, astroglial, and germ layer markers through qPCR. Neuronal and glial protein expression were analyzed by immunocytochemistry.
ResultsBoth compounds alone and in combination induced differentiation in MSCs with significant gene expression of neuronal markers i.e. neuron specific enolase (NSE), nestin, microtubule-associated protein 2 (MAP2), neurofilament light chain (Nefl) and Tau, and astroglial marker i.e. glial fibrillary acidic protein (GFAP). AP treated group also showed significant upregulation of endodermal and mesodermal markers indicating transition of ectoderm towards the other two germ layers.
ConclusionThis study concludes that AP and TQ potentially differentiate MSCs into neuronal and astroglial lineages. However, AP treated group followed germ layer transition. Expression of neuronal as well as glial markers indicate that the differentiated neurons are at the neuroprogenitor stage and can be potential candidates for cellular therapeutics against neurodegenerative disorders.
Keywords: Neurodegeneration, Neuronal differentiation, Neuroprogenitor, GFAP, Germ layers -
Pages 155-169Introduction
Cell-based models play an important role in understanding the pathophysiology and etiology of auditory disorders. For the auditory system, models have primarily focused on restoring inner and outer hair cells. However, they have largely underrepresented the surrounding structures and cells that support the function of the hair cells.
MethodsIn this article, we will review recent advancements in the evolution of cell-based models of auditory disorders in their progression towards three dimensional (3D) models and organoids that more closely mimic the pathophysiology in vivo.
ResultsWith the elucidation of the molecular targets and transcription factors required to generate diverse cell lines of the components of inner ear, research is starting to progress from two dimensional (2D) models to a greater 3D approach. Of note, the 3D models of the inner ear, including organoids, are relatively new and emerging in the field. As 3D models of the inner ear continue to evolve in complexity, their role in modeling disease will grow as they bridge the gap between cell culture and in vivo models.
ConclusionUsing 3D cell models to understand the etiology and molecular mechanisms underlying auditory disorders holds great potential for developing more targeted and effective novel therapeutics.
Keywords: Cell-based model, Auditory system, Spiral ganglion neuron, Organoid model, 3D model, Stem cell -
Pages 171-174
The cholinergic anti-inflammatory pathway (CAP) first described by Wang et al, 2003 has contemporary interest arising from the COVID-19 pandemic. While tobacco smoking has been considered an aggravating factor in the severity of COVID-19 infections, it has been suggested by some that the nicotine derived from tobacco could lessen the severity of COVID-19 infections. This spotlight briefly describes the CAP and its potential role as a therapeutic target for the treatment of COVID-19 infections using vagus nerve stimulation or selective alpha7 nicotinic acetylcholine receptor agonists.
-
Pages 175-177
The impact of gut as the origin of different disorders has led to the "gut-origin concept" of diseases. The gut microbiome regulates host defenses against viral infections, thus dysbiosis can play a major role in triggering the cascade of inflammation and causing immune imbalances in COVID-19 patients. It appears that gut microbial signature in COVID-19 patients can be used as a potential diagnostic, therapeutic, and even a prognostic marker. Personalized nutrition therapy can be used by profiling the gut microbiota of individual patients and specialized probiotics/synbiotics to modify gut dysbiosis. Hence, improving overall immune responses can be recommended in these patients.
Keywords: Gut microbiota, Probiotics, Gut lung axis, COVID-19